This workshop is designed for participants with command-line knowledge. You will need to be able to ssh into a remote machine, navigate the directory structure and scp files from a remote computer to your local computer.
Description
What is the influence of genotype (intrinsic) and environment (extrinsic) on anemone-associated bacterial communities?
Data: Illumina MiSeq v3 paired-end (2 × 300 bp) reads (FASTQ).
Tools: QIIME 2
Pipeline:
- Section 1: Importing, cleaning and quality control of the data
- Section 2: Taxonomic Analysis
- Section 3: Building a phylogenetic tree
- Section 4: Basic visualisations and statistics
- Section 5: Exporting data for further analysis in R
- Section 6: Extra Information
Learning Objectives
At the end of this introductory workshop, you will:
- Take raw data from a sequencing facility and end with publication quality graphics and statistics
- Answer the question What is the influence of genotype (intrinsic) and environment (extrinsic) on anemone-associated bacterial communities?
Tutorial Layout
- There is a
Table of contentson the top of the page which can be used to easily navigate through the tutorial by clicking the relevant section.
These black coloured boxes are code blocks. The rectangular boxes in the top
right hand corner of this code block/black box can be used to copy the code to
the clipboard.Coloured boxes like these with > on the far right hand side, can be clicked to reveal the contents.
REVEALED!
Attention: Pay attention to the information in these boxes.
Important information, hints and tips.
Requirements and Preparation
Important
Attendees are required to use their own laptop computers.
At least one week before the workshop, if required, participants should install the software below. This should provide sufficient time for participants to liaise with their own IT support should they encounter any IT problems.
Mode of Delivery
This workshop will be run on the SAIAB lab417 server.
Information on how to access the lab417 server is explained in Introduction to the Unix shell and High Performance Computing.
You will need to use a Google Chrome or Mozilla Firefox web browser to view files in QIIME2 View.
Required Data
- No additional data needs to be downloaded for this workshop - it is all located on the lab417 server. FASTQs are located in the directory
raw_dataand a metadata (metadata.tsv) file has also been provided. - If you wish to analyse the data independently at a later stage, it can be downloaded from here. This zipped folder contains both the FASTQs and associated metadata file.
- If you are running this tutorial independently, you can also access the classifier that has been trained specifically for this data from here.
Background
What is the influence of genotype (intrinsic) and environment (extrinsic) on anemone-associated bacterial communities?
The Players
- Exaiptasia diaphana - a shallow-water, marine anemone that is often used in research as a model organism for corals. In this experiment, two genotypes (AIMS1 and AIMS4) of E. diaphana were grown in each of two different environments:
- sterile seawater OR
- unfiltered control seawater
- The anemone-associated bacterial communities or microbiome - these bacteria live on, or within E. diaphana, and likely consist of a combination of commensals, transients, and long-term stable members, and combined with their host, form a mutually beneficial, stable symbiosis.
The Study
The anemone microbiome contributes to the overall health of this complex system and can evolve in tandem with the anemone host. In this data set we are looking at the impact of intrinsic and extrinsic factors on anemone microbiome composition. After three weeks in either sterile or control seawater (environment), anemones were homogenized and DNA was extracted. There are 23 samples in this data set - 5 from each anemone treatment combination (2 genotypes x 2 environments) and 3 DNA extraction blanks as controls. This data is a subset from a larger experiment.
Model Organism: Exaiptasia diaphana - a shallow-water marine anemone used as a coral research model
Experimental Duration: 3 weeks in respective environments
Analysis: DNA extraction and microbiome composition analysis
Dungan AM, van Oppen MJH, and Blackall LL (2021) Short-Term Exposure to Sterile Seawater Reduces Bacterial Community Diversity in the Sea Anemone, Exaiptasia diaphana. Front. Mar. Sci. 7:599314. doi:10.3389/fmars.2020.599314 [Full Text].
QIIME 2 Analysis Platform
Attention
The version used in this workshop is qiime2-amplicon-2024.10. Other versions of QIIME2 may result in minor differences in results.
Quantitative Insights Into Microbial Ecology 2 (QIIME 2™) is a next-generation microbiome bioinformatics platform that is extensible, free, open source, and community developed. It allows researchers to:
- Automatically track analyses with decentralised data provenance
- Interactively explore data with beautiful visualisations
- Easily share results without QIIME 2 installed
- Plugin-based system — researchers can add in tools as they wish
Qiime2 is built around a few key principles:
Core Architecture
1. Artifact System (.qza files)
- Everything in QIIME2 is stored as "artifacts" - special files that contain:
- Your actual data (sequences, tables, trees, etc.)
- Metadata about how the data was created
- Complete provenance tracking (every step that created this data)
2. Plugin Architecture
- QIIME2 is modular - different tools are "plugins"
- Each plugin has specific "actions" (commands you can run)
- Examples:
q2-dada2,q2-feature-classifier,q2-diversity
3. Semantic Types
- Data has specific "types" that QIIME2 understands
- Examples:
SampleData[PairedEndSequencesWithQuality],FeatureTable[Frequency],Phylogeny[Rooted] - This prevents you from using the wrong data type with incompatible methods
Basic QIIME2 Workflow
Raw sequences (.fastq)
↓ [Import]
Sequence artifact (.qza)
↓ [Quality control + Denoising]
Feature table + Representative sequences (.qza)
↓ [Taxonomic classification]
Feature table + Taxonomy table (.qza)
↓ [Diversity analysis]
Alpha/Beta diversity results (.qza/.qzv)
Taxonomic Analysis in Detail
Taxonomic analysis answers the question: "What organisms are in my samples?
The Challenge
You have DNA sequences that look like this:
AGGATTAGATACCCTGGTAGTCCACGCCGTAAACGATGTCGATTTGGA...
You need to know: "Is this from E. coli? Bacteroides? Something unknown?"
How QIIME2 Solves This
Step 1: Prepare a Classifier
A classifier is like a "reference library" that has been trained to recognize sequences:
qiime feature-classifier classify-sklearn \
--i-classifier my-classifier.qza \
--i-reads representative-seqs.qza \
--o-classification taxonomy.qza
- Takes known sequences from databases (SILVA, Greengenes, etc.)
- Uses machine learning (Naive Bayes) to learn patterns
- Creates a "trained model" that can classify new sequences
Viewing QIIME2 Visualisations
Attention
In order to use QIIME2 View to visualise your files, you will need to use a Google Chrome or Mozilla Firefox web browser (not in private browsing). For more information, click here.
As this workshop is being run on a remote lab417 server, you will need to download the visual files (*.qzv) to your local computer and view them in QIIME2 View (q2view).
Attention
We will be doing this step multiple times throughout this workshop to view visualisation files as they are generated.
Alternatively, you have QIIME2 installed and are running it on your own computer, you can use qiime tools view to view the results from the command line (e.g. qiime tools view filename.qzv). qiime tools view opens a browser window with your visualization loaded in it. When you are done, you can close the browser window and press ctrl-c on the keyboard to terminate the command.
Section 1: Importing, cleaning and quality control of the data
Initial Set up on lab417 server
Using screen command
To ensure that commands continue to run should you get disconnected from your lab417 server, we'll run a screen session.
Starting a screen session on lab417
On lab417, to start a screen session called workshop, type
screen -S workshopIf you get disconnected from your lab417 Instance, follow the instructions here to resume your session.
Should your SSH session on lab417 terminate, once you log back in to your lab417 instance, list running sessions/screens:
screen -lsIf it says (Detached) next to the workshop session in the list, reattach to workshop by:
screen -r workshopIf it says (Attached) next to the workshop session in the list, you can access workshop which is already attached by:
screen -r -d workshop/Detaching or Terminating a screen session.
To detach from workshop, type ctrl-a ctrl-d while inside the workshop session. Follow the information on the screen and select 1 for Screen mode.
ctrl-d while inside the workshop session.
Activate Qiime2 conda environment
Activate the Qiime2 environment with this command:
source /usr/local/bin/qiime2-activate
Verify that it is working with this command:
qiime infoSymbolic links to workshop data
Data for this workshop is stored in a central location (/opt/courses/qiime/data_files) on the lab417 file system that we will be using. We will use symbolic links (ln -s) to point to it.
cd
ln -s /opt/courses/qiime/data_files/raw_data raw_data
ln -s /opt/courses/qiime/data_files/metadata.tsv metadata.tsv
ln -s /opt/courses/qiime/data_files/silva_138_16s_v5v6_classifier_2021-4.qza silva_138_16s_v5v6_classifier_2021-4.qzaImport data
These samples were sequenced on a single Illumina MiSeq run using v3 (2 × 300 bp) reagents at the Walter and Eliza Hall Institute (WEHI), Melbourne, Australia.
Data from WEHI came as paired-end, demultiplexed, unzipped *.fastq files with adapters still attached. Following the QIIME2 importing tutorial, this is the Casava One Eight format. The files have been renamed to satisfy the Casava format as SampleID_FWDXX-REVXX_L001_R[1 or 2]_001.fastq e.g. CTRLA_Fwd04-Rev25_L001_1_001.fastq.gz. The files were then zipped (.gzip).
Here, the data files (two per sample i.e. forward and reverse reads 1 and 2 respectively) will be imported and exported as a single QIIME 2 artefact file. These samples are already demultiplexed (i.e. sequences from each sample have been written to separate files), so a metadata file is not initially required.
Note
To check the input syntax for any QIIME2 command, enter the command, followed by --help e.g. qiime tools import --help
Start by making a new directory analysis to store all the output files from this tutorial. In addition, we will create a subdirectory called seqs to store the exported sequences.
cd
mkdir -p analysis/seqsRun the command to import the raw data located in the directory raw_data and export it to a single QIIME 2 artefact file, combined.qza.
qiime tools import \
--type 'SampleData[PairedEndSequencesWithQuality]' \
--input-path raw_data \
--input-format CasavaOneEightSingleLanePerSampleDirFmt \
--output-path analysis/seqs/combined.qzaRemove primers
Important
Remember to ask your sequencing facility if the raw data you get has the primers attached - they may have already been removed.
These sequences still have the primers attached - they need to be removed (using cutadapt) before denoising.
qiime cutadapt trim-paired \
--i-demultiplexed-sequences analysis/seqs/combined.qza \
--p-front-f AGGATTAGATACCCTGGTA \
--p-front-r CRRCACGAGCTGACGAC \
--p-error-rate 0.20 \
--output-dir analysis/seqs_trimmed_data \
--verboseAttention
The primers specified (784f and 1492r for the bacterial 16S rRNA gene) correspond to this specific experiment - they will likely not work for your own data analyses.
Create and interpret sequence quality data
Create a viewable summary file so the data quality can be checked. Viewing the quality plots generated here helps determine trim settings.
Things to look for:
- Where does the median quality drop below 30?
- Do any of the samples have only a few sequences e.g. <1000?: If so, you may want to omit them from the analysis later on in R.
Create a subdirectory in analysis called visualisations to store all files that we will visualise in one place.
mkdir analysis/visualisations
qiime demux summarize \
--i-data analysis/seqs_trimmed_data/trimmed_sequences.qza \
--o-visualization analysis/visualisations/trimmed_sequences.qzv
Copy analysis/visualisations/trimmed_sequences.qzv to your local computer and view in QIIME 2 view (q2view).
Visualisations: Read quality and demux output
click to view the trimmed_sequences.qzv file in QIIME 2 View!
Make sure to switch between the “Overview” and “Interactive Quality Plot” tabs in the top left hand corner. Click and drag on plot to zoom in. Double click to zoom back out to full size. Hover over a box to see the parametric seven-number summary of the quality scores at the corresponding position.
Things to look for:
- Overview tab shows summary statistics of your trimmed sequencesand. What to look for:
- Sequence retention rate: You want to keep 70-90% of your original sequences
- Length consistency: For amplicon data, sequences should be similar lengths
- No dramatic drops: Large losses suggest overly aggressive trimming
- Interactive Quality Plot shows per-position nucleotide quality scores. What to look for:
- 5' end quality: Should be high (Q30+)
- 3' end degradation: Natural quality drop is normal
- Sudden quality drops: May indicate adapter contamination
- Overall quality: Most positions should be >Q20
Denoising the data
Trimmed data sequences are now quality assessed using the dada2 plugin within QIIME2. dada2 denoises data by modelling and correcting Illumina-sequenced amplicon errors, and infers exact amplicon sequence variants (ASVs), resolving differences of as little as 1 nucleotide. Its workflow consists of filtering, de-replication, reference‐free chimera detection, and paired‐end reads merging, resulting in a feature or ASV table.
Note
This step may long time to run (i.e. hours), depending on files sizes and computational power.
Remember to adjust p-trunc-len-f and p-trunc-len-r values according to your own data.
Based on your assessment of the quality plots from the visual inspection of the trimmed_sequences.qzv file generated in the previous step, what values would you select for p-trunc-len-f and p-trunc-len-r in the command below?
Hint: At what base pair does the median quality drop below 30?
Answer
For version qiime2-2021.8: p-trunc-len-f 211 and p-trunc-len-r 167. Other QIIME2 versions may slightly differ. Upload your trimmed_sequences.qzv file to QIIME2 view, change to the “Interactive Quality Plot” tab and zoom in on the plots to find the relevant base pairs for the QIIME2 version you are using.
The specified output directory must not pre-exist.
The qiime dada2 denoise-paired command is the core processing step for paired-end amplicon sequencing data in QIIME2. It performs quality filtering, error correction, denoising, read merging, and chimera removal to produce high-quality Amplicon Sequence Variants (ASVs).
DADA2 performs several critical steps in sequence:
- Quality filtering - Removes low-quality reads
- Error model learning - Learns sequencing error patterns
- Denoising - Corrects sequencing errors
- Paired-end merging - Combines forward and reverse reads
- Chimera detection - Removes chimeric sequences
- ASV generation - Creates exact sequence variants
qiime dada2 denoise-paired \
--i-demultiplexed-seqs analysis/seqs_trimmed_data/trimmed_sequences.qza \
--p-trunc-len-f 211 \
--p-trunc-len-r 167 \
--p-n-threads 0 \
--output-dir analysis/dada2out \
--verbose
The output from this step is summarized with the next few commands below.
Generate summary files
A metadata file is required which provides the key to gaining biological insight from your data. The filemetadata.tsv is provided in the home directory of your lab417 instance. This spreadsheet has already been verified using the plugin for Google Sheets, keemei.
The qiime metadata tabulate command on the file denoising_stats.qza creates a visualization showing how many reads were retained or lost at each step of the DADA2 denoising process. This is crucial quality control information.
Things to look for:
- Percentage of input passed filter - Percentage reads remaining after quality filtering based on
--p-max-ee, --p-trunc-len, --p-trim-leftparameters. Good retention: >70-80% of reads. - Percentage of input merged - Percentage of forward and reverse reads with sufficient overlap. Typical retention: 85-95% of merged reads.
- Percentage of input non-chimeric - Percentage of reads that does not have chimeric sequences (PCR artifacts). Typical retention: 85-95% of merged reads.
This visualization is essential for validating your DADA2 parameters and ensuring you haven't over- or under-filtered your data, directly impacting the quality and reliability of downstream diversity and taxonomic analyses.
qiime metadata tabulate \
--m-input-file analysis/dada2out/denoising_stats.qza \
--o-visualization analysis/visualisations/16s_denoising_stats.qzv \
--verbose
Copy analysis/visualisations/16s_denoising_stats.qzv to your local computer and view in QIIME 2 View (q2view).
Visualisations: Denoising Stats
The qiime feature-table summarize command is one of the most important commands in QIIME2 analysis. It generates comprehensive statistics about your feature table (ASV/OTU table) and is essential for making informed decisions about downstream analysis parameters. It creates a detailed summary of your feature table - the matrix that shows how many times each ASV (Amplicon Sequence Variant) appears in each sample.
Things to look for in the output:
- Overview Tab - Provides high-level statistics about your dataset. Number of samples in your dataset, number of unique ASVs detected
- Interactive Sample Detail Tab - Shows per-sample statistics
- Interactive Feature Detail Tab - Shows per-ASV statistics
Healthy Dataset Indicators include:
- even distribution of sequences across samples
- reasonable number of ASVs (hundreds to thousands)
- most samples have > 1,000 sequences and
- gradual decline in ASV abundance (not all equal)
qiime feature-table summarize \
--i-table analysis/dada2out/table.qza \
--m-sample-metadata-file metadata.tsv \
--o-visualization analysis/visualisations/16s_table.qzv \
--verbose
Copy analysis/visualisations/16s_table.qzv to your local computer and view in QIIME 2 View (q2view).
Visualisations: Feature/ASV summary
Click to view the 16s_table.qzv.qzv file in QIIME 2 View!
Make sure to switch between the “Overview” and “Feature Detail” tabs in the top left hand corner.

The qiime feature-table tabulate-seqs command creates a searchable, interactive table that displays the actual DNA sequences for each feature (ASV) in your dataset. It's essential for examining, validating, and understanding the biological content of your features.
Things to look for:
It takes your representative sequences (the actual DNA sequences for each ASV) and creates an interactive visualization where you can:
- View the actual DNA sequences
- Search and filter sequences
- Link sequences to external databases
- Examine sequence properties
qiime feature-table tabulate-seqs \
--i-data analysis/dada2out/representative_sequences.qza \
--o-visualization analysis/visualisations/16s_representative_seqs.qzv \
--verbose
Copy analysis/visualisations/16s_representative_seqs.qzv to your local computer and view in QIIME 2 View (q2view).
Visualisations: Representative Sequences
Good sequences should be appropriate length for your primer pair and start/end at expected positions. Show sequence diversity appropriate for your samples and not contain obvious primer or adapter sequences and match expected gene region (16S, ITS, etc.)
Section 2: Taxonomic Analysis
Assign taxonomy
Here we will classify each identical read or Amplicon Sequence Variant (ASV) to the highest resolution based on a database. Common databases for bacterial datasets are Greengenes, SILVA, Ribosomal Database Project, or Genome Taxonomy Database. See Porter and Hajibabaei, 2020 for a review of different classifiers for metabarcoding research. The classifier chosen is dependent upon:
- Previously published data in a field
- The target region of interest
- The number of reference sequences for your organism in the database and how recently that database was updated.
A classifier in QIIME2 is a pre-trained machine learning model that assigns taxonomic classifications to DNA sequences (specifically ASVs/OTUs) by comparing them against a reference database. It's one of the most crucial components of microbiome analysis. The classifier takes your unknown DNA sequences and answers the question: "What organism does this sequence come from?"
A classifier has already been trained for you for the V5V6 region of the bacterial 16S rRNA gene using the SILVA database. The next step will take a while to run. The output directory cannot previously exist.
n_jobs = 1 This runs the script using all available cores
Note
The classifier used here is only appropriate for the specific 16S rRNA region that this data represents. You will need to train your own classifier for your own data. For more information about training your own classifier, see Section 6: Extra Information.
STOP - Workshop participants only
Due to time limitations in a workshop setting, please do NOT run the qiime feature-classifier classify-sklearn command below. You will need to access a pre-computed classification.qza file that this command generates by running the following: cd
mkdir analysis/taxonomy
cp /opt/courses/qiime/data_files/classification.qza analysis/taxonomy/.
If you have accidentally run the command below, ctrl-c will terminate it.
qiime feature-classifier classify-sklearn \
--i-classifier silva_138_16s_v5v6_classifier_2021-4.qza \
--i-reads analysis/dada2out/representative_sequences.qza \
--p-n-jobs 1 \
--output-dir analysis/taxonomy \
--verbose
Warning
This step often runs out of memory on full datasets. Some options are to change the number of cores you are using (adjust --p-n-jobs) or add --p-reads-per-batch 10000 and try again. The QIIME 2 forum has many threads regarding this issue so always check there was well.
Generate a viewable summary file of the taxonomic assignments.
The qiime metadata tabulate command creates interactive, searchable tables from QIIME2 metadata and statistical results based on the input files. It's one of the most versatile visualization commands, essential for examining sample metadata, denoising statistics, diversity metrics, and other tabular data. This command is your data exploration hub - essential for quality control, results interpretation, and preparing data for downstream statistical analysis!
Here for example we generate a viewable summary file of the taxonomic assignments
qiime metadata tabulate \
--m-input-file analysis/taxonomy/classification.qza \
--o-visualization analysis/visualisations/taxonomy.qzv \
--verbose
Copy analysis/visualisations/taxonomy.qzv to your local computer and view in QIIME 2 View (q2view).
Visualisations: Taxonomy
Things to look for:
Key Information in Your Taxonomy Table:
- Feature ID Column
- Unique identifiers for each Amplicon Sequence Variant
- Links to your feature table and representative sequences
- Usually numbered sequentially or hashed
- Taxon Column
- k__ = Kingdom
- p__ = Phylum
- c__ = Class
- o__ = Order
- f__ = Family
- g__ = Genus
- s__ = Species
- Confidence Column
- 0.999 = 99.9% confidence (excellent)
- 0.856 = 85.6% confidence (good)
- 0.652 = 65.2% confidence (moderate)
- 0.423 = 42.3% confidence (poor)
Example:k__Bacteria;p__Firmicutes;c__Bacilli;o__Lactobacillales;f__Streptococcaceae
- Classification stops at family level
Implications: Genus/species unknown or low confidence
Filtering
Filter out reads classified as mitochondria and chloroplast. Unassigned ASVs are retained.
According to QIIME developer Nicholas Bokulich, low abundance filtering (i.e. removing ASVs containing very few sequences) is not necessary under the ASV model.
qiime taxa filter-table \
--i-table analysis/dada2out/table.qza \
--i-taxonomy analysis/taxonomy/classification.qza \
--p-exclude Mitochondria,Chloroplast \
--o-filtered-table analysis/taxonomy/16s_table_filtered.qza \
--verbose
Generate a viewable summary file of the new table to see the effect of filtering.
qiime feature-table summarize \
--i-table analysis/taxonomy/16s_table_filtered.qza \
--m-sample-metadata-file metadata.tsv \
--o-visualization analysis/visualisations/16s_table_filtered.qzv \
--verbose
Copy analysis/visualisations/16s_table_filtered.qzv to your local computer and view in QIIME 2 View (q2view).
Visualisations: 16s_table_filtered
Things to look for:
The 16s_table_filtered.qzv file is one of the most important visualizations in your entire QIIME2 analysis. It tells you about the quality and characteristics of your data after filtering. Here's what to examine:
- Tab 1: "Overview" - Your Dataset Summary
- Number of samples: Should match your expected sample count
- Number of features: Total unique ASVs in your dataset
- Total frequency: Total sequence reads across all samples
- Tab 2: "Interactive Sample Detail" - Per-Sample Analysis
- Frequency per sample (sequence counts): Shows how many sequences each sample has. Look for the histogram and statistics.
- Rarefaction Depth Decision:
- The median is often a good rarefaction depth.
- Don't go below the 1st quartile (you'll lose too many samples)
- Don't go above the 3rd quartile (you'll lose too much data per sample)
- Tab 3: "Feature Detail" - Per-ASV Analysis
- Feature frequency distribution: Shows how common/rare your ASVs are. Most ASVs should be rare (typical microbiome pattern)
- Abundance patterns:
- Most abundant features: Should dominate
- Medium abundance: Moderate numbers
- Rare features: Many features with few reads
- Key Decisions Based on This File
- Rarefaction Depth for Diversity Analysis:
Use the sample frequency data to choose
(Example: if median = 5,583, use this for diversity analysis)qiime diversity core-metrics-phylogenetic --p-sampling-depth 5583 - Sample Filtering Decisions: Identify problematic samples
(Exclude samples with very low counts) - Data Quality Assessment:
Good: Even distribution, reasonable sample sizes
Problematic: Extreme outliers, many failed samples
For this anemone microbiome data, look for:
Extraction blanks: Should have very low counts
Treatment groups: Similar count distributions?
Biological replicates: Consistent within treatments?
Section 3: Build a phylogenetic tree
The next step does the following:
- Perform an alignment on the representative sequences.
- Mask sites in the alignment that are not phylogenetically informative.
- Generate a phylogenetic tree.
- Apply mid-point rooting to the tree.
A phylogenetic tree is necessary for any analyses that incorporates information on the relative relatedness of community members, by incorporating phylogenetic distances between observed organisms in the computation. This would include any beta-diversity analyses and visualisations from a weighted or unweighted Unifrac distance matrix.
mkdir analysis/tree
analysis/phylogeny
Use one thread only (which is the default action) so that identical results can be produced if rerun.
qiime phylogeny align-to-tree-mafft-fasttree \
--i-sequences analysis/dada2out/representative_sequences.qza \
--o-alignment analysis/tree/aligned_16s_representative_seqs.qza \
--o-masked-alignment analysis/tree/masked_aligned_16s_representative_seqs.qza \
--o-tree analysis/tree/16s_unrooted_tree.qza \
--o-rooted-tree analysis/tree/16s_rooted_tree.qza \
--p-n-threads 1 \
--verbose
Section 4: Basic visualisations and statistics
ASV relative abundance bar charts
Create bar charts to compare the relative abundance of ASVs across samples.
qiime taxa barplot \
--i-table analysis/taxonomy/16s_table_filtered.qza \
--i-taxonomy analysis/taxonomy/classification.qza \
--m-metadata-file metadata.tsv \
--o-visualization analysis/visualisations/barchart.qzv \
--verbose
Copy analysis/visualisations/barchart.qzv to your local computer and view in QIIME 2 View (q2view). Try selecting different taxonomic levels and metadata-based sample sorting.
Visualisations: Taxonomy Barplots
Click to view the 16s_table_filtered.qzv file in QIIME 2 View!
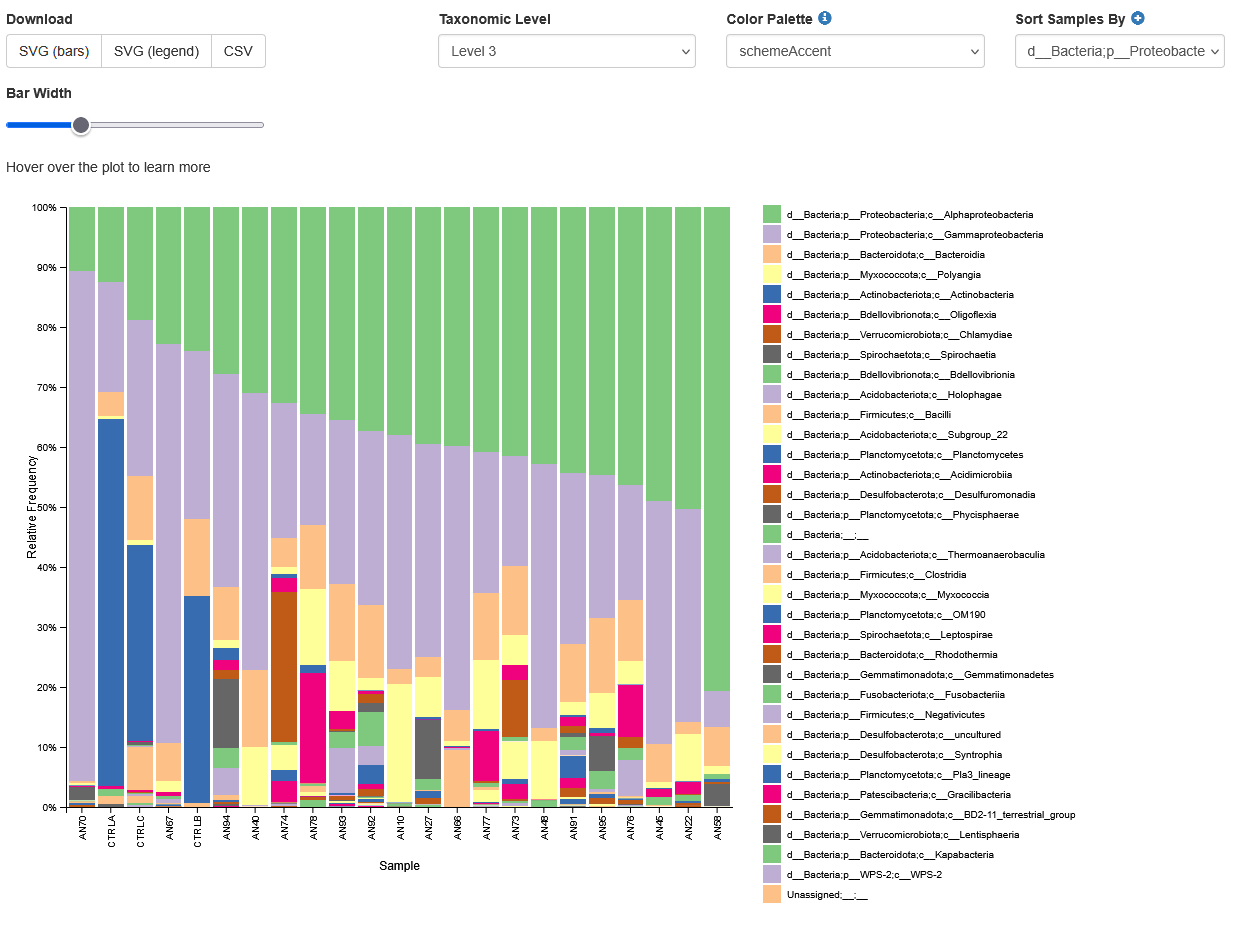
Increase the “Bar Width”, select Index in “Sort Samples By” drop-down menu and explore the resulting barplots by changing the levels in the “Change Taxonomic Level” dropdown menu (Select Level 1, then Level 3, and then Level 5 for example).
You should see something like the image below showing Level 3 taxons
Rarefaction curves
Generate rarefaction curves to determine whether the samples have been sequenced deeply enough to capture all the community members.
Things to look for:
- Do the curves for each sample plateau? If they don't, the samples haven't been sequenced deeply enough to capture the full diversity of the bacterial communities.
- At what sequencing depth (x-axis) do your curves plateau? This value will be important for downstream analyses.
Note
The value that you provide for –p-max-depth should be determined by reviewing the “Frequency per sample” information presented in the 16s_table_filtered.qzv file that was created above. In general, choosing a value that is somewhere around the median frequency seems to work well, but you may want to increase that value if the lines in the resulting rarefaction plot don’t appear to be leveling out, or decrease that value if you seem to be losing many of your samples due to low total frequencies closer to the minimum sampling depth than the maximum sampling depth.
qiime diversity alpha-rarefaction \
--i-table analysis/taxonomy/16s_table_filtered.qza \
--i-phylogeny analysis/tree/16s_rooted_tree.qza \
--p-max-depth 9062 \
--m-metadata-file metadata.tsv \
--o-visualization analysis/visualisations/16s_alpha_rarefaction.qzv \
--verbose
Copy analysis/visualisations/16s_alpha_rarefaction.qzv to your local computer and view in QIIME 2 View (q2view).
Visualisations: Rarefaction
Click to view the 16s_table_filtered.qzv file in QIIME 2 View!
Select “Genotype” in the “Sample Metadata Column”:
Alpha and beta diversity analysis
The following is taken directly from the Moving Pictures tutorial and adapted for this data set. QIIME 2’s diversity analyses are available through the q2-diversity plugin, which supports computing alpha- and beta- diversity metrics, applying related statistical tests, and generating interactive visualisations. We’ll first apply the core-metrics-phylogenetic method, which rarefies a FeatureTable[Frequency] to a user-specified depth, computes several alpha- and beta- diversity metrics, and generates principle coordinates analysis (PCoA) plots using Emperor for each of the beta diversity metrics.
The metrics computed by default are:
- Alpha diversity (operate on a single sample (i.e. within sample diversity)).
- Shannon’s diversity index (a quantitative measure of community richness)
- Observed OTUs (a qualitative measure of community richness)
- Faith’s Phylogenetic Diversity (a qualitative measure of community richness that incorporates phylogenetic relationships between the features)
- Evenness (or Pielou’s Evenness; a measure of community evenness)
- Beta diversity (operate on a pair of samples (i.e. between sample diversity)).
- Jaccard distance (a qualitative measure of community dissimilarity)
- Bray-Curtis distance (a quantitative measure of community dissimilarity)
- unweighted UniFrac distance (a qualitative measure of community dissimilarity that incorporates phylogenetic relationships between the features)
- weighted UniFrac distance (a quantitative measure of community dissimilarity that incorporates phylogenetic relationships between the features)
An important parameter that needs to be provided to this script is --p-sampling-depth, which is the even sampling (i.e. rarefaction) depth that was determined above. As most diversity metrics are sensitive to different sampling depths across different samples, this script will randomly subsample the counts from each sample to the value provided for this parameter. For example, if --p-sampling-depth 500 is provided, this step will subsample the counts in each sample without replacement, so that each sample in the resulting table has a total count of 500. If the total count for any sample(s) are smaller than this value, those samples will be excluded from the diversity analysis. Choosing this value is tricky. We recommend making your choice by reviewing the information presented in the 16s_table_filtered.qzv file that was created above. Choose a value that is as high as possible (so more sequences per sample are retained), while excluding as few samples as possible.
qiime diversity core-metrics-phylogenetic \
--i-phylogeny analysis/tree/16s_rooted_tree.qza \
--i-table analysis/taxonomy/16s_table_filtered.qza \
--p-sampling-depth 5583 \
--m-metadata-file metadata.tsv \
--output-dir analysis/diversity_metrics
Copy the .qzv files created from the above command into the visualisations subdirectory.
cp analysis/diversity_metrics/*.qzv analysis/visualisations
To view the differences between sample composition using unweighted UniFrac in ordination space, copy analysis/visualisations/unweighted_unifrac_emperor.qzv to your local computer and view in QIIME 2 View (q2view).
Visualisations: Unweighted UniFrac Emperor Ordination
Click to view the unweighted_unifrac_emperor.qzv file in QIIME 2 View!
On q2view, select the “Color” tab, choose “Environment” under the “Select a Color Category” dropdown menu, then select the “Shape” tab and choose “Genotype” under the “Select a Shape Category” dropdown menu.

Other metrics calculated above can also be viewed by copying the files in analysis/visualisations/ to your local computer and view in QIIME 2 View (q2view).
Next, we’ll test for associations between categorical metadata columns and alpha diversity data. We’ll do that here for the Faith Phylogenetic Diversity (a measure of community richness) and Evenness metrics.
qiime diversity alpha-group-significance \
--i-alpha-diversity analysis/diversity_metrics/faith_pd_vector.qza \
--m-metadata-file metadata.tsv \
--o-visualization analysis/visualisations/faith-pd-group-significance.qzv
Copy analysis/visualisations/faith-pd-group-significance.qzv to your local computer and view in QIIME 2 View (q2view).
Visualisations: Faith Phylogenetic Diversity output
Click to view the faith-pd-group-significance.qzv file in QIIME 2 View!
Select “Environment” under the “Column” dropdown menu.
What is Faith's Phylogenetic Diversity (Faith's PD)?
Faith's PD measures the total branch length of a phylogenetic tree that spans all taxa present in a sample. Unlike simple species richness (counting number of species), Faith's PD accounts for how evolutionarily distinct the species are from each other.
- Higher Faith PD = More phylogenetically diverse community. Often associated with healthy, stable ecosystems.
- Lower Faith PD = Less phylogenetically diverse community (more closely related species).
qiime diversity alpha-group-significance \
--i-alpha-diversity analysis/diversity_metrics/evenness_vector.qza \
--m-metadata-file metadata.tsv \
--o-visualization analysis/visualisations/evenness-group-significance.qzv
Copy analysis/visualisations/evenness-group-significance.qzv to your local computer and view in QIIME 2 View (q2view).
Visualisations: Evenness output
Click to view the evenness-group-significance.qzv file in QIIME 2 View!
Select “Genotype” under the “Column” dropdown menu.
What is Evenness?
Evenness measures how equally abundant the different species are in a community, independent of the total number of species (richness).
- High evenness (→1.0): All species have similar abundances (balanced community)
- Low evenness (→0.0): Few species dominate, many are rare (unbalanced community)
Finally, we’ll analyse sample composition in the context of categorical metadata using a permutational multivariate analysis of variance (PERMANOVA, first described in Anderson (2001)) test using the beta-group-significance command. The following commands will test whether distances between samples within a group, such as samples from the same genotype, are more similar to each other then they are to samples from the other groups. If you call this command with the --p-pairwise parameter, as we’ll do here, it will also perform pairwise tests that will allow you to determine which specific pairs of groups (e.g., AIMS1 and AIMS4) differ from one another, if any. This command can be slow to run, especially when passing --p-pairwise, since it is based on permutation tests. So, unlike the previous commands, we’ll run beta-group-significance on specific columns of metadata that we’re interested in exploring, rather than all metadata columns to which it is applicable. Here we’ll apply this to our unweighted UniFrac distances, using two sample metadata columns, as follows.
qiime diversity beta-group-significance \
--i-distance-matrix analysis/diversity_metrics/unweighted_unifrac_distance_matrix.qza \
--m-metadata-file metadata.tsv \
--m-metadata-column Genotype \
--o-visualization analysis/visualisations/unweighted-unifrac-genotype-significance.qzv \
--p-pairwise
Copy analysis/visualisations/unweighted-unifrac-genotype-significance.qzv to your local computer and view in QIIME 2 View (q2view).
Visualisations: Genotype significance output
Click to view the unweighted-unifrac-genotype-significance.qzv file in QIIME 2 View!
Select “Genotype” under the “Column” dropdown menu.
What is Unweighted UniFrac?
Unweighted UniFrac genotype significance analysis tests whether microbial community composition differs significantly between genotype groups using phylogenetic beta diversity.
It uses evolutionary distances between species and only considers presence/absence (not abundance). Focuses on which species are present, not how abundant they are.
qiime diversity beta-group-significance \
--i-distance-matrix analysis/diversity_metrics/unweighted_unifrac_distance_matrix.qza \
--m-metadata-file metadata.tsv \
--m-metadata-column Environment \
--o-visualization analysis/visualisations/unweighted-unifrac-environment-significance.qzv \
--p-pairwise
Copy analysis/visualisations/unweighted-unifrac-environment-significance.qzv to your local computer and view in QIIME 2 View (q2view).
Visualisations: Environmental significance output
Section 5: Exporting data for further analysis in R
You need to export your ASV table, taxonomy table, and tree file for analyses in R. Many file formats can be accepted.
Export unrooted tree as .nwk format as required for the R package phyloseq.
qiime tools export \
--input-path analysis/tree/16s_unrooted_tree.qza \
--output-path analysis/exportCreate a BIOM table with taxonomy annotations. A FeatureTable[Frequency] artefact will be exported as a BIOM v2.1.0 formatted file.
qiime tools export \
--input-path analysis/taxonomy/16s_table_filtered.qza \
--output-path analysis/export
Then export BIOM to TSV
biom convert \
-i analysis/export/feature-table.biom \
-o analysis/export/feature-table.tsv \
--to-tsv
Export Taxonomy as TSV
qiime tools export \
--input-path analysis/taxonomy/classification.qza \
--output-path analysis/export
Delete the header lines of the .tsv files
sed '1d' analysis/export/taxonomy.tsv > analysis/export/taxonomy_noHeader.tsv
sed '1d' analysis/export/feature-table.tsv > analysis/export/feature-table_noHeader.tsv
Some packages require your data to be in a consistent order, i.e. the order of your ASVs in the taxonomy table rows to be the same order of ASVs in the columns of your ASV table. It’s recommended to clean up your taxonomy file. You can have blank spots where the level of classification was not completely resolved.
Section 6: Extra Information¶
Note
This section contains information on how to train the classifier for analysing your own data. This will NOT be covered in the workshop.
Train SILVA v138 classifier for 16S/18S rRNA gene marker sequences.
SILVA provides comprehensive, quality checked and regularly updated datasets of aligned small (16S/18S, SSU) and large subunit (23S/28S, LSU) ribosomal RNA (rRNA) sequences for all three domains of life (Bacteria, Archaea and Eukarya). The newest version of the SILVA database (v138) can be trained to classify marker gene sequences originating from the 16S/18S rRNA gene. Reference files silva-138-99-seqs.qza and silva-138-99-tax.qza were downloaded from SILVA and imported to get the artefact files. You can download both these files from here.
Reads for the region of interest are first extracted. You will need to input your forward and reverse primer sequences. See QIIME2 documentation for more information.
qiime feature-classifier extract-reads \
--i-sequences silva-138-99-seqs.qza \
--p-f-primer FORWARD_PRIMER_SEQUENCE \
--p-r-primer REVERSE_PRIMER_SEQUENCE \
--o-reads silva_138_marker_gene.qza \
--verbose
The classifier is then trained using a naive Bayes algorithm. See QIIME2 documentation for more information.
qiime feature-classifier fit-classifier-naive-bayes \
--i-reference-reads silva_138_marker_gene.qza \
--i-reference-taxonomy silva-138-99-tax.qza \
--o-classifier silva_138_marker_gene_classifier.qza \
--verbose